
Vassilis Koutkias
@vkoutkias
Principal Researcher, Institute of Applied Biosciences, Centre for Research & Technology Hellas, Greece

OpenPVSignal
The newly developed OpenPVSignal model offers improved search, share, and reuse of signal information.
Signal communication is a key pharmacovigilance activity. When a signal is detected, the respective information is disseminated through periodic newsletters and targeted commentaries, which can be electronic or paper-based. For example, the bi-monthly WHO Pharmaceuticals Newsletter devotes a section to signals. Many pharmacovigilance centres, such as the Netherlands Pharmacovigilance Centre Lareb, publish signal information on their websites. But, while this information is publicly available, its unstructured format does not optimally support systematic access, search, and data interlinking.
Typically, signals contain a title referring to the adverse reaction and the respective drugs; the authors; a summary or introduction; evidence supporting the signal, such as individual case safety reports or the medical literature; a conclusion; and the respective bibliographic references.
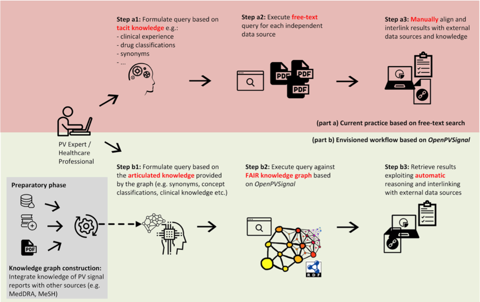
Currently, pharmacovigilance professionals looking for signal information have to search across diverse, unstructured signal sources, then aggregate the information of interest based on their tacit knowledge and personal experience. This is a time-consuming procedure with multiple manual steps, heavily dependent on the technical skills of each professional.
To address that problem, a group of biomedical informatics researchers established by the Institute of Applied Biosciences at the Centre for Research and Technology-Hellas (CERTH) has developed OpenPVSignal, a novel knowledge model through which signals can be annotated and enhanced with metadata. The idea of OpenPVSignal arose from discussions among the group and pharmacovigilance professionals about their routine practice, which identified the need to improve communication and search of signal information.
OpenPVSignal makes two key contributions. First, it enables signal information to be published according to the FAIR (findable, accessible, interoperable, and re-usable) data principles. And second, it improves the dissemination of signal information through data interlinking and automatic processing.
In particular, OpenPVSignal supports a well-defined processing workflow enabling the construction of a “knowledge graph”, which can be queried by pharmacovigilance professionals. The knowledge graph is instantiated using the content of the unstructured text within signals and is linked to other relevant data through references to external terminologies and thesauri, such as the Anatomical Therapeutic Chemical (ATC) Classification System, the Medical Dictionary for Regulatory Activities (MedDRA®), Medical Subject Headings (MeSH), and others. This way, OpenPVSignal supports efficient data integration and browsing, minimises end-user effort, prevents errors and missing information links, and enables automatic inferencing using linked data.
“OpenPVSignal supports a well-defined processing workflow enabling the construction of a knowledge graph.”
OpenPVSignal can be useful to drug monitoring organisations, pharmacovigilance departments in hospitals, pharmaceutical companies, and contract research organisations. The aim is not to substitute current approaches for signal communication with OpenPVSignal, but rather to offer a way to further enrich signal information through automatic data interlinking and processing. Following the FAIR data principles, OpenPVSignal reinforces transparency and reuse of such important drug safety information.
In a recent publication, the developers demonstrated the use of OpenPVSignal through case studies, elaborating on three different sources of pharmacovigilance signals: signals contained in the WHO Pharmaceuticals Newsletter, signals from the Netherlands Pharmacovigilance Centre Lareb, and announcements contained in the FDA Drug Safety Communication. These case studies illustrated the value of OpenPVSignal in automatically interlinking data of clinical importance among diverse signals, which was difficult to identify manually. The study also successfully presented the evaluation of OpenPVSignal in terms of its FAIR compliance.
The team followed a transparent process for developing OpenPVSignal, collaborating with pharmacovigilance professionals to help both define and validate the model.
To encourage further feedback, the software is available as an open-access resource on GitHub.
Currently, a set of user-friendly tools is under development to enable pharmacovigilance professionals to easily instantiate the OpenPVSignal model and search it. The developers also plan to conduct a more thorough validation by elaborating on signals published by other organisations and accommodate potential extensions of the model. And, given the large number of signals that have been published, the developers are considering appropriate text processing methods to extract signal information and populate the OpenPVSignal model accordingly.
Overall, it appears that OpenPVSignal can be the basis for an advanced signal communication mechanism and improved, more efficient pharmacovigilance workflows.
“It is important to share information about signals of adverse drug reactions on a global level. OpenPVSignal can facilitate this by making signal information more easily searchable and, thus, more accessible.”
– Linda Härmark, Head of Innovation and Projects, Netherlands Pharmacovigilance Centre Lareb, The Netherlands.
“The new ontology proposed by V. Koutkias and his colleagues is a major advance for our practice. The FAIR principles are perfectly in resonance with our deontology, such as traceability and quality standards. The interconnection of heterogeneous data is a major advance, as is the semantic enrichment, because of the complexity of the terminologies and thesauri available. The accessibility and interoperability ensure that all pharmacovigilance teams worldwide could use it. And the added value of interlinking and reasoning as demonstrated in the article opens huge perspectives for us. We can’t wait for the next steps of the OpenPVSignal.”
– Agnès Lillo-Le Louët, Head, Centre Régional de Pharmacovigilance, Hôpital Européen Georges-Pompidou, AP-HP, Paris, France.
“The approach described in the article is a very appealing one. Publishing signal information in a standardised format, as suggested, would make it possible to automatically request and assemble information from all available sources globally without the need to pool all this information in one place. The information should be retrieved and at hand – when and where it is needed.”
– Magnus Wallberg, Technology Evangelist, Uppsala Monitoring Centre, Uppsala, Sweden.
The following people assisted the author with this article:
South Africa’s pharmacovigilance system has been evolving for over a decade with UMC’s data management system supporting them every step of the way.
02 September 2024
Egypt's swift detection of vincristine-associated necrotising infections demonstrates how effective pharmacovigilance can identify risks and protect patients globally.
08 May 2025
The safety of orphan drugs is hard to monitor as they target rare diseases afflicting small patient populations. However, an initiative led by the SFDA aims to improve this.
12 August 2024