
Lucie Gattepaille
@LucieGattepa
Data Science Team Manager, UMC

Could natural language processing unlock powerful new methods in pharmacovigilance science? See how a journey through space could lead to a new dimension in case report analysis.
How do you tell whether two individual case safety reports (ICSRs) are similar? This is the question I began asking myself early in my employment at Uppsala Monitoring Centre (UMC). At the time, I had just come out of a two-month project for improving our duplicate detection algorithm, and some aspects of it had left me a bit unsatisfied. The gist of how our algorithm works is that it looks at different important fields of two reports (for example, age, gender, country of origin, reported reactions, reported drugs) and computes a score that rewards matching information and penalises mismatches, weighing in how frequent each piece of information is in the first place. When it came to the reported reactions, this approach just felt too rough.
Consider how large MedDRA is: if two independent persons review a given case, it is not at all certain that they would choose to describe the observed reactions using the exact same set of MedDRA terms. If I choose “blood pressure increased” and you choose “hypertension”, are we really describing different things? To circumvent this, we could of course soften the strict “match/no match” criterion by going up the MedDRA hierarchy and allow for some reward when two terms share the same high level term (HLT) instead of penalising the mismatch, but this would not solve our problem above, since the term I chose and the one you chose are from different system organ classes (SOCs) – a classic problem when dealing with lab results and their associated diagnoses. Conversely, there are terms in MedDRA that share an HLT but that we would not want to match. For example, we would not want to match a case of “foot amputation” with a case of “hand amputation” when trying to identify duplicates.
“We needed a way to take relatedness of terms into account in a much more nuanced way than MedDRA allows”
We needed something else. We needed a way to take relatedness of terms into account in a much more nuanced way than MedDRA allows. Obviously, even the whole staff of UMC would never be enough for manually scoring all pairs of terms for relatedness, so whatever solution we could come up with, it would have to be data-driven. Thankfully, UMC has over the years gathered a team of smart data scientists and all it took was to draw a parallel between our current problem and a very similar one in a quite different domain, namely Natural Language Processing (NLP).
In NLP, people have been struggling for decades with the problem of representing words in a clever way, a way that would somehow model the meaning of words. How do you tell whether two sentences are similar? Well, they are similar if the words they are made of carry similar meanings. To a linguist (though maybe not a meeting planner) the words “Monday” and “Tuesday”, however different, are similar in meaning. From an NLP perspective, they are in most cases entirely interchangeable. What NLP researchers then tried to achieve was to develop methods of producing numeric representations of words so that similar words get similar representations. With these methods, algorithms for NLP could be built upon these representations to become “meaning-aware”. Distributional semantic methods arose out of these efforts.
“You shall know a word by the company it keeps”
Distributional semantic methods are best summarised by this quote: “You shall know a word by the company it keeps” (John Rupert Firth, British linguist, 1957). The central idea is that if you see two words surrounded by similar contexts – that is, with a high degree of interchangeability – then these words are likely to carry a similar meaning. The approach gained great popularity in the early 2010s when a distributional semantic method called word2vec was published, leading to a great boost in performance of algorithms in many kinds of classical NLP tasks (such as part-of-speech tagging, machine translation, question-answering, and document classification).
In essence, what word2vec does is to model every word as a vector (think simply of a list of numeric values) and learn each vector component with machine learning by trying to predict centre words in context windows, using massive amounts of text. Details of the method are probably uninteresting for the non-technical crowd, but the essential concept to understand is that the method creates a space (imagine a 3D-space for example) where similar words have similar coordinates. In a region somewhere in the space, you might find “Monday”, “Tuesday”, and all the other days of the week, while in another region you might find “cat”, “dog”, and other kinds of pets.
From here, addressing our problem of automatically finding similarities across case reports becomes quite straightforward. After all, just like a sentence provides a context for words, an ICSR provides a context for both reported drugs and reported preferred terms (PTs). So, we applied word2vec to more than 16 million VigiBase reports, creating a novel “space of meaning” for both drugs and events. Preliminary quantitative and qualitative analyses showed that the resulting vectors were indeed meaningful, as illustrated by the tables below, which provide lists of nearest neighbours in the space for different concepts of interest. Now, if I choose “Blood pressure increased” and you choose “Hypertension”, the new representations show that we are not so far from each other.
“In the way people report drugs and reactions in VigiBase, there is meaning in these concepts – and it is truly mind blowing”
|
Blood pressure systolic increased |
|
Blood pressure increased |
|
Blood pressure fluctuation |
|
Blood pressure diastolic increased |
|
Blood pressure abnormal |
|
Anaphylactic shock |
|
Anaphylactoid reaction |
|
Laryngeal oedema |
|
Bronchospasm |
|
Type I hypersensitivity |
|
Bupropion |
|
Sertraline |
|
Venlafaxine |
|
Paroxetine |
|
Buspirone |
|
Certolizumab pegol |
|
Golimumab |
|
Vedolizumab |
|
Tocilizumab |
|
Abatacept |
All these new vector representations create a space of meaning, built in an entirely data-driven way based on co-reporting patterns present in VigiBase. In theory, only a human with the proper expertise can make sense of each reported term and each reported drug on a given ICSR – in other words we can’t expect a machine to make any sense of them, because that is not what VigiBase has been designed for. Nevertheless, in the way people report drugs and reactions in VigiBase, there is meaning in these concepts. And it is truly mind blowing.
As much as being data-driven is a blessing, it is also in some respects its own curse. At the core, it is difficult to understand why the concepts get the coordinates they do, why X is the nearest neighbour of Y instead of Z. Evaluating the quality and soundness of the space is also a challenge because there is no clear truth to compare it to.
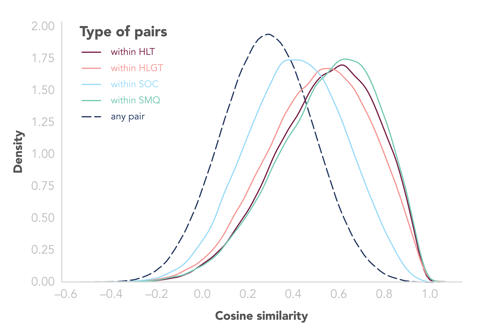
In my attempt to validate the vector representations, I used the MedDRA and ATC hierarchies, based on the assumption that two PTs or two drugs that share an ancestor in the hierarchy should, in general, have more similar coordinates, and that similarity should decrease as the ancestor gets less specific (for example, we would expect two PTs sharing an HLT to be closer in the space than two sharing only one SOC or none at all). And I did see the expected pattern: overall, similarity between drugs or PTs decreased as we relaxed the constraints on their shared ancestries in the hierarchies (see the chart below).
However, we see that there is still a large overlap between distributions, indicating that the relationships retained in the space are more complex and certainly go beyond the MedDRA and ATC hierarchies. We also have to keep in mind that, for the algorithm to be able to build good vector representation, it requires data, so representations of rare PTs and drugs are likely to be noisy and less reliable. But we believe that by using these vector representations in all kinds of pharmacovigilance tasks, we will be able to prove their potential and their usefulness.
Using vector representations, we now have a way (albeit not perfect) to automatically quantify relatedness between important concepts such as drugs and PT. This means we are now in a much better position to compare reports with one another, as was my original intention. Duplicate detection, report clustering, case series expansion – these are tasks that could, already now, benefit from this new way of representing drugs and PTs. But that’s not all. For example, we could also improve disproportionality analysis by harnessing the power of entire neighbourhoods of terms. Suggestions for coding alternatives during data entry could be integrated into VigiFlow. Since the new representations of drugs and PTs are numeric, they can be the basis for insightful visualisations, like ADR profiles for drugs, which could be integrated in VigiLyze in the future. By computing distances to labelled ADRs, we could speed up signal detection, identifying drug–reaction combinations that are captured in basic form in the label information. And thanks to their numeric form and semantic content, the representations are likely to be much more powerful inputs for machine learning algorithms and statistical methods.
As shown with all these examples, I truly believe that the paradigm shift in representing drugs and PTs introduced by this new space has the potential to reshape many areas of pharmacovigilance science and spur novel advancements in the field. We have now at our disposal an entire space that begs to be explored, so let’s board our spaceships and boldly go where no one has gone before!
If you want to know more details about the method we used, please check our scientific article online or contact us.
Read more:
Want to hear more?
Tune in to UMC's Drug Safety Matters podcast for an audio version of this article and an interview with Lucie Gattepaille about her work and its implications for drug safety research. Or listen to the episode right here...
AI is already reshaping drug safety. Niklas Norén explores how to harness its power responsibly in this two-part episode.
26 November 2025
Discover how researchers are addressing crucial challenges in pregnancy-related pharmacovigilance to better protect both mothers and their unborn children.
03 April 2025
AI is already reshaping drug safety. Niklas Norén explores how to harness its power responsibly in this two-part episode.
22 October 2025